TransAtlasDB
An Informatics system for Transcriptome Analysis Data
TransAtlasDB Quick Start Guide
TransAtlasDB is a sophisticated bioinformatics system, which incorporates both the Relational Database and NoSQL Database system.
The relational database, MySQL, enforces data assurance and integrity and the NoSQL database system, FastBit, ensures rapid performance for querying big data.
Hence, both database systems are required to be installed and added to the systems’ or users’ executable path.
BEFORE YOU BEGIN
First, we need to download appropriate databases and required Perl modules from the Usage & Downloads page, before proceeding to the next section.
Assume that we have downloaded TransAtlasDB toolkit and unpacked the TAR package using
You will see that the directory contains several Perl programs with .pl suffix.
IMPORTANT: Make sure the TransAtlasDB folder is in your desired permanent location before proceeding, preferably the
NOTE: If you already added the transatlasdb path into your system executable path, then typing
INSTALLATION
The TransAtlasDB database system and necessary components need to be installed to a local disk using INSTALL-tad.pL and user must have root privileges to install.
IMPORTANT: TransAtlasDB installation requires superuser (root) privileges.
Only the
# Syntax for installing TransAtlasDB using defaults and mysql password as 'mysql-password'.
sudo perl transatlasdb-location/INSTALL-tad.pL -password mysql-password
Otherwise, arguments such as the mysql username
# Syntax for installing TransAtlasDB specifying username as 'mysql-username' and databasename as 'mysql-databasename'.
sudo perl INSTALL-tad.pL -p mysql-password -username mysql-username -databasename mysql-databasename
The NoSQL folder-name
# Syntax for installing TransAtlasDB specifying username as 'mysql-username' and databasename as 'mysql-databasename' with nosql folder 'nosqlfolder'.
sudo perl INSTALL-tad.pL -p mysql-password -u mysql-username -d mysql-databasename -fastbitname nosqlfolder
Connecting to the database using connect-tad.pL
The installational module needs to be carried out once per local disk to prevent database access conflict. However, if such conflict arises user settings can be viewed and/or corrected for using connect-tad.pL.
# Syntax for viewing user installation details.
connect-tad.pL
# Syntax for setting existing user installation details. Parameters are the same as the INSTALL-tad.pL script.
connect-tad.pL -p mysql-password -u mysql-username -d mysql-databasename -fastbitname nosqlfolder
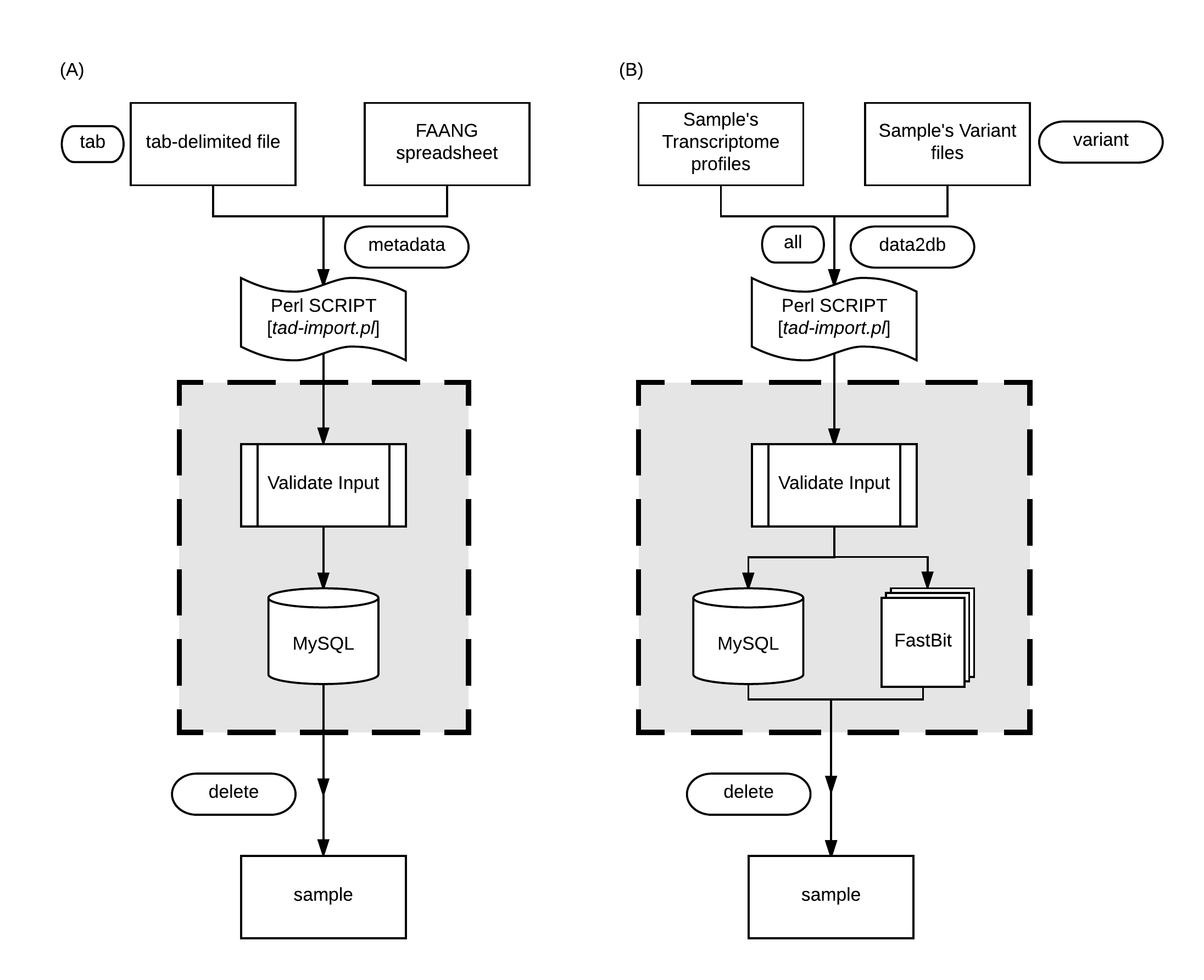
DATA IMPORT
Transcriptome analysis data can be imported using tad-import.pl.
Importing Samples Metadata
The samples information, commonly known as, samples metadata consists of the relevant details to uniquely identify each specimen used for RNAseq.
The samples metadata can be imported via
# Syntax for importing the example FAANG metadata provided.
tad-import.pl -metadata example/metadata/FAANG/FAANG_GGA_UD.xlsx
# The second option to import a tab-delimited file using the example provided.
tad-import.pl -metadata -t example/metadata/TEMPLATE/metadata_GGA_UD.txt
Importing Transcriptome Analysis Data
Transcriptome analysis results can be inserted using
# Syntax for importing all RNASeq expression and variant results in for sample 'GGA_UD_1004'.
tad-import.pl -data2db -all example/sample_sxt/GGA_UD_1004
# Syntax for importing RNASeq expression profiling information in for sample 'GGA_UD_1004'.
tad-import.pl -data2db -gene example/sample_sxt/GGA_UD_1004
# Syntax for importing RNASeq variant results in for sample 'GGA_UD_1004'.
tad-import.pl -data2db -variant example/sample_sxt/GGA_UD_1004
Importing Variant Annotation information
The variant functional annotations predicted from either VEP or ANNOVAR can also be imported using additional flags
# Syntax for importing RNASeq variant results with VEP annotations for sample 'GGA_UD_1004'.
tad-import.pl -data2db -variant -vep example/sample_sxt/GGA_UD_1014
# Syntax for importing RNASeq data with ANNOVAR annotations for sample 'GGA_UD_1004'.
tad-import.pl -data2db -all example/sample_sxt/GGA_UD_1004 -annovar
NOTE: The analysis data should be stored in a single folder for each sample and the folder-name must be the same sample name as represented in the samples metadata. A typical folder directory structure is shown here.
DATA RETRIEVAL
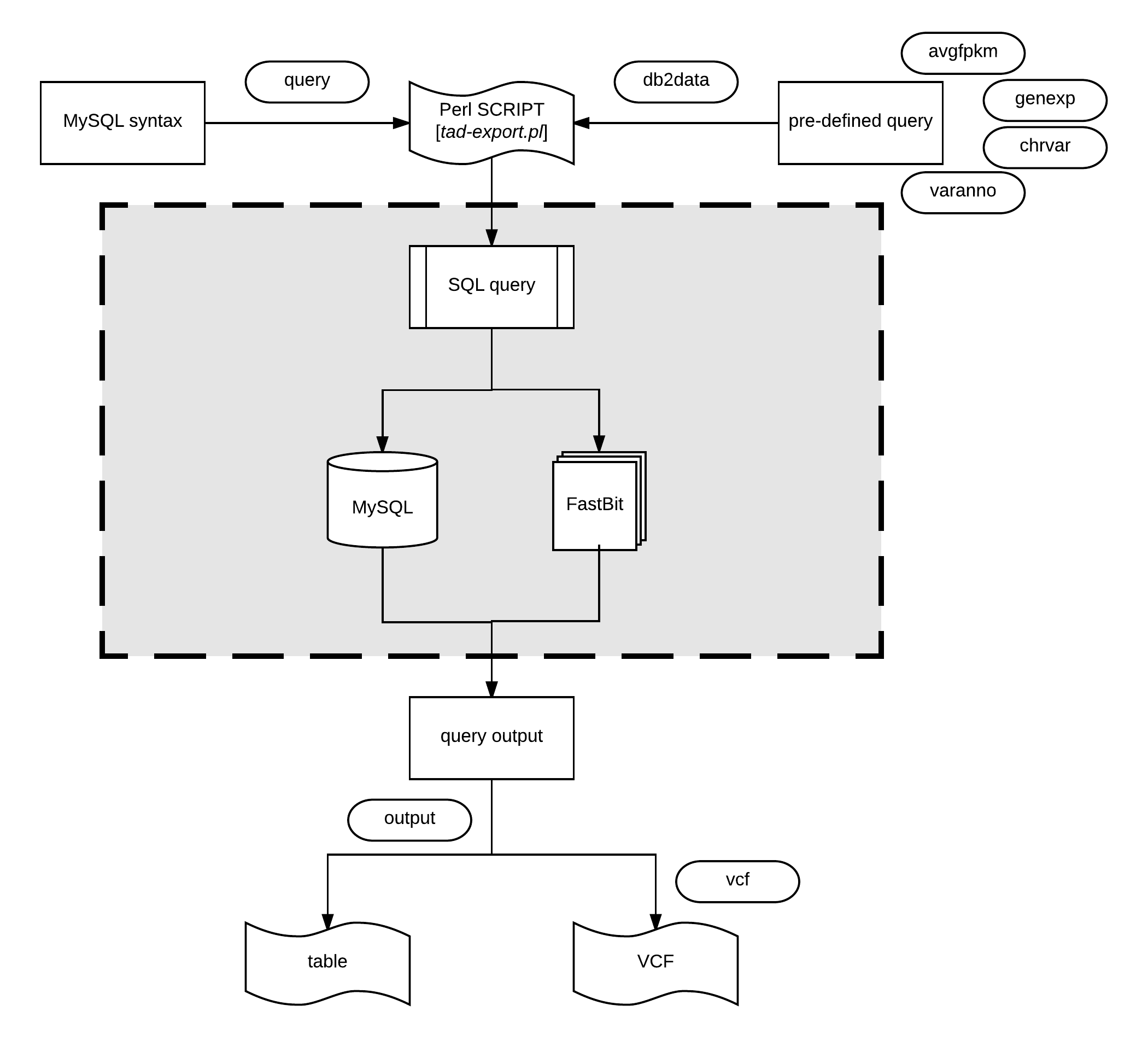
Transcriptome analysis data previously stored can be retrieved using tad-export.pl.
NOTE: Analysis data can also be retrieved using the web interface.
The export module offers two methods of extracting data from the database; one by performing data manipulation language (DML) SQL statements using
Performing DML Statements
Users can execute direct SQL statements via
More information on writing Select statements to FastBit can be viewed here.
For instance, executing 'show tables' will retrieve all the rows currently in the database, which can be stored as a tab-delimited file.
# Syntax to view all the tables in the database.
tad-export.pl -query 'show tables'
# Syntax to retrieve all the rows in the Sample table and store the results in a tab-delimited file 'output.txt'.
tad-export.pl -query 'select * from Sample' -output output.txt
# Syntax on nosql database, to view the first ten rows in the gene-information folder.
tad-export.pl -nosql gene-information -query 'select genename,organism,tissue,fpkm,tpm where 1=1 limit 10'
Executing Pre-defined Queries of Research Interest
The pre-defined user statements can be accessed via
- Average expression values of specified genes organized by the different tissues.
Average expression values via --avgfpkm flag
View the average, maximum and minimum expression (FPKM) values of specified genes given species and/or tissue(s) of interest.# Syntax to view the expression values of genes 'MST' and 'GDF' for the 'Gallus gallus' specie in the database.
tad-export.pl -db2data -avgfpkm -gene 'MST,GDF' -species 'Gallus gallus'
# To export prior syntax as a tab-delimited file 'output.txt'.
tad-export.pl -db2data -avgfpkm -gene 'MST,GDF' -species 'Gallus gallus' -o output.txt
- Gene expression profiles across the different samples of the same organism. Specific samples can be selected.
Gene expression profiles via --genexp flag
View expression values of individual genes per sample. This is done to easily compare expression profiles of the different samples.# Syntax to view all the genes of all samples in the database for 'Gallus gallus' organism.
tad-export.pl -db2data -genexp -species 'Gallus gallus'
# To export prior syntax as a tab-delimited file 'output.txt'.
tad-export.pl -db2data -genexp -species 'Gallus gallus' -o output.txt
# Syntax to view expression profiles of genes 'OPTN' and 'GDF' for samples 'GGA_UD_1004'.
tad-export.pl -db2data -genexp -species 'Gallus gallus' -gene 'OPTN,GDF' -sample 'GGA_UD_1004' -o output.txt
- Variant distribution of all, or selected chromosomes for individual samples in the database.
Variant distribution of chromosomes via --chrvar flag
Summary counts of the different variant types per chromosome for each sample.# Syntax to view chromosome variant counts for all samples in the database for 'Gallus gallus' organism.
tad-export.pl -db2data -chrvar -species 'Gallus gallus'
# To export prior syntax as a tab-delimited file 'output.txt'.
tad-export.pl -db2data -chrvar -species 'Gallus gallus' -o output.txt
# Syntax to view variant count fo expression profiles of chromosomes 'chr1, chr2, chr3' for all samples.
tad-export.pl -db2data -chrvar -species 'Gallus gallus' -chromosome 'chr1,chr2,chr3' -o output.txt
# Syntax to view variant count fo expression profiles of chromosomes 'chr1, chr2, chr3' for sample 'GGA_UD_1004'.
tad-export.pl -db2data -chrvar -species 'Gallus gallus' -chromosome 'chr1,chr2,chr3' -sample 'GGA_UD_1004' -o output.txt
- Variants and predicted functional annotations found in the organism or selected genes or chromosomes.
Variants and functional annotations via --varanno flag
View gene-associated variants and chromosomal region-associated variants with respective annotation information. and annotation information.# Syntax to view associated variants with respective annotation information for 'OPTN' gene in 'Gallus gallus' organism.
tad-export.pl -db2data -varanno -gene 'OPTN' -species 'Gallus gallus'
# Syntax to view associated variants with respective annotation for chromosomes 'chr1,chr4,chr7' in 'Gallus gallus' organism.
tad-export.pl -db2data -varanno -chromosome 'chr1, chr4, chr7' -species 'Gallus gallus'
# Syntax to view associated variants with respective annotation for chr1:50000-900000 in 'Gallus gallus' organism.
tad-export.pl -db2data -varanno -chromosome 'chr1' -region 50000-900000 -species 'Gallus gallus'
# To export prior syntax as a tab-delimited file 'output.txt'.
tad-export.pl -db2data -varanno -chromosome 'chr1' -region 50000-900000 -species 'Gallus gallus' -o output.txt
# To export prior syntax as a vcf file 'output.vcf'.
tad-export.pl -db2data -varanno -chromosome 'chr1' -region 50000-900000 -species 'Gallus gallus' -o -vcf output.vcf
ADDITIONAL PROGRAMS
Interacting with TransAtlasDB using tad-interact.pl
If uncertain on how to proceed with the export module, the interaction module provides an easy-to-use menu-driven interface.
The menu offers seven choices of exploratory research interest and provides a detailed description of what can be done from the module.
# Syntax to begin the interaction module.
tad-interact.plNOTE: The interaction module only outputs a small subset of results and further instructions on how to export the complete results will be displayed.
Deleting Data
Sample results can only be imported once to ensure data integrity, nonetheless, previously imported data can be cautiously deleted using
# Syntax for deleting sample 'GGA_UD_1004' from the database.
tad-import.pl -delete GGA_UD_1004
WEB ENVIRONMENT
Biologist can benefit from using the web interface to interact with the analysis data previously stored using the data import module.
The web interface is designed into five major sections:
About
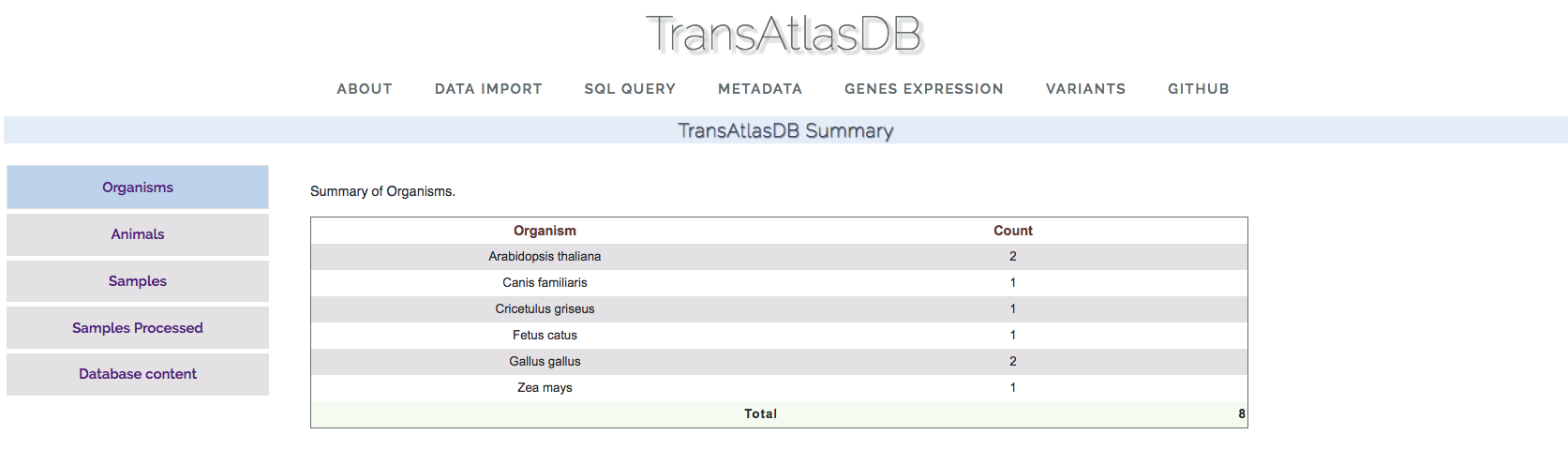
The about page gives the summary statistics of the samples data imported in the database, and consist of four parts.- Organisms:
This sections displays the organisms and total number of samples present in the database.
- Animals:
This section displays the number of animals for each organism in the database.

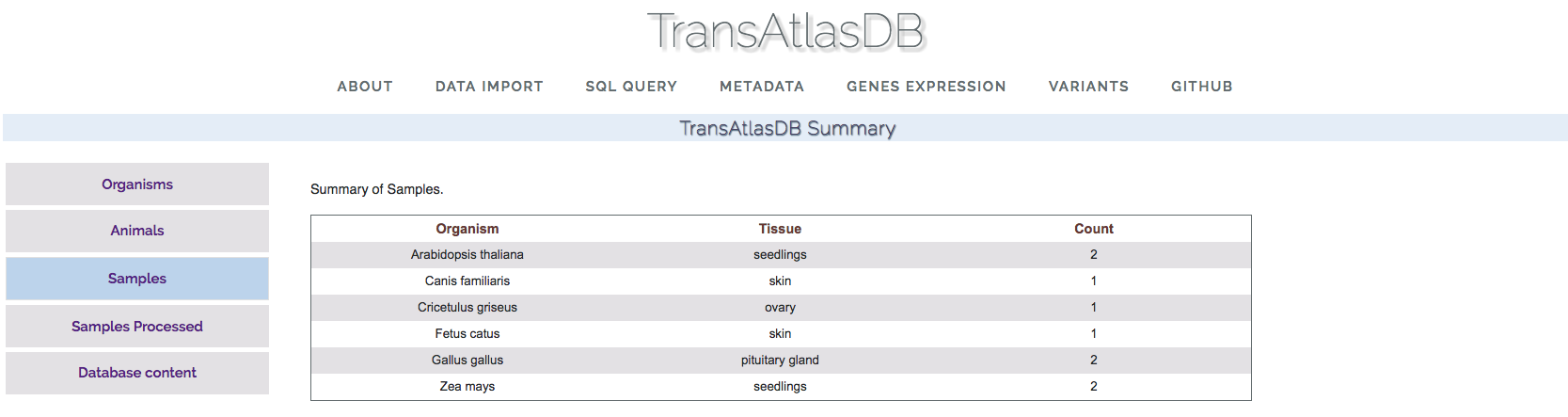
- Samples:
This section displays the organisms and total number of samples for each tissue in the database.
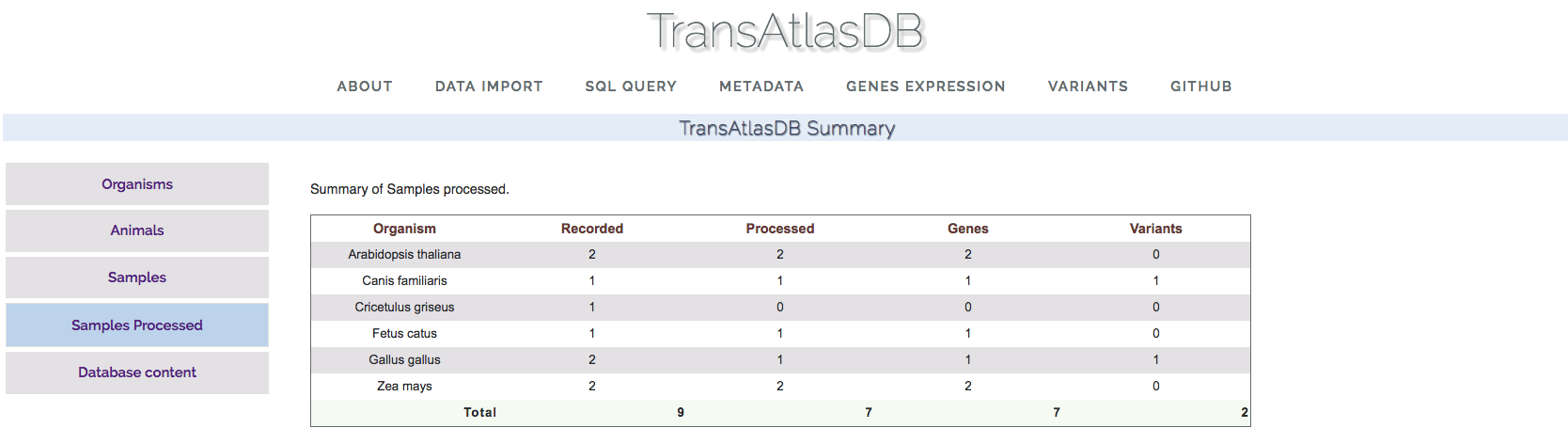
- Samples Processed:
This section displays the summary counts of the samples already archived in the database.
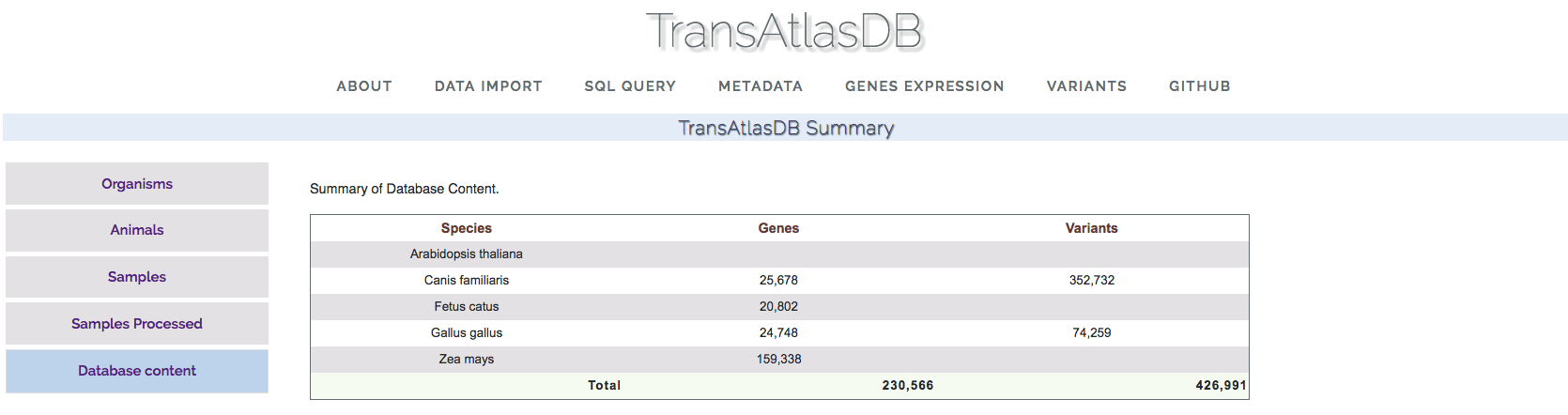
- Database Content:
This section displays the summary count of the variants and genes present in the database.
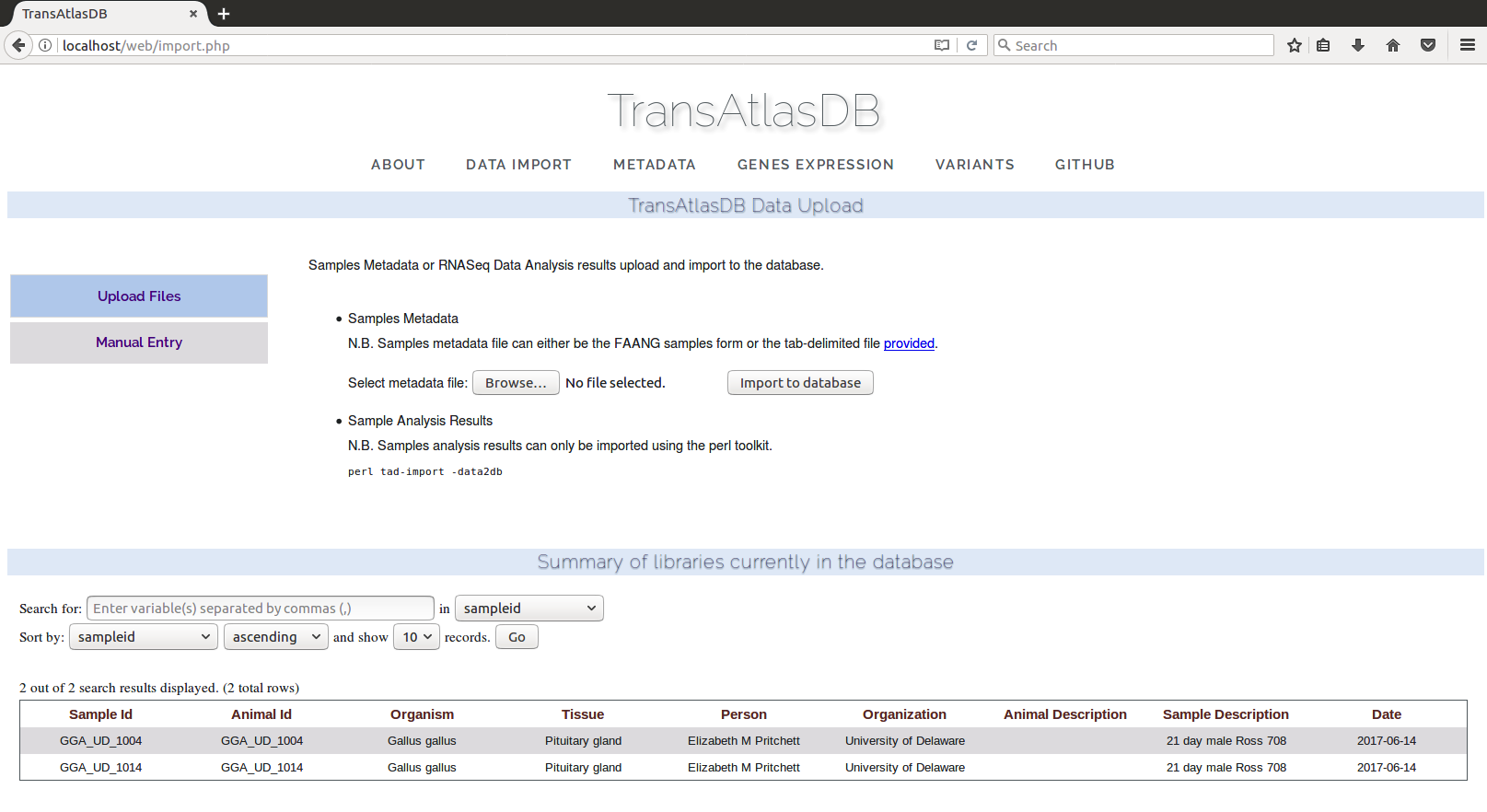
Data Import
The data import page provides two methods of importing the samples metadata. Storing the large data files such as the gene expression profiling and variants analysis results can only be done using the Perl toolkit as explained above.- File Upload:
Sample metadata are uploaded from a sample file, either using the sample tab-delimited template or FAANG spreadsheet.
NOTE: At this point, only sample metadata files can be uploaded via the web interface. RNAseq analyses data files should be imported through the command-line toolkit.
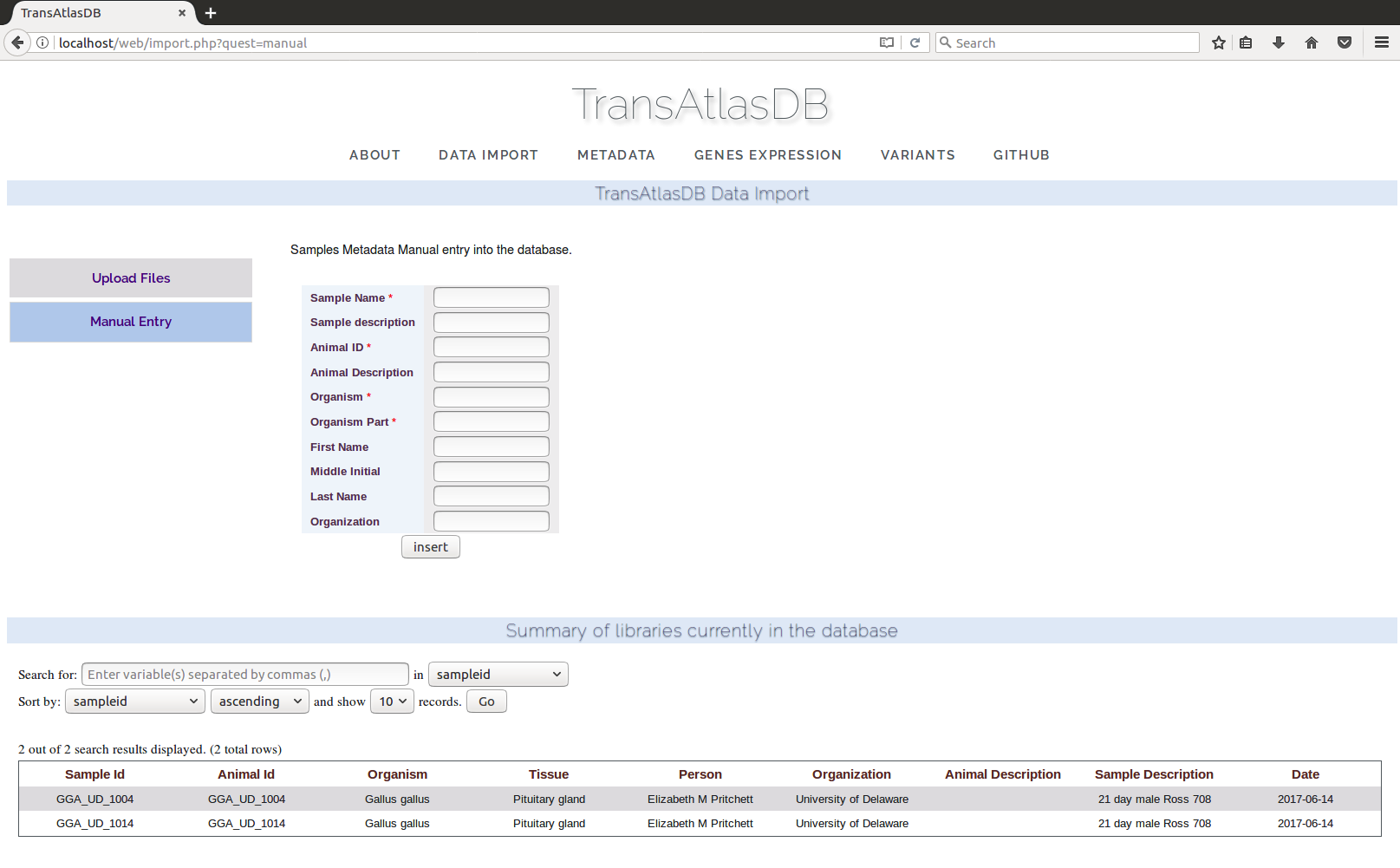
- Manual Entry:
Samples metadata can also be inserted by manually entering the information.
Metadata
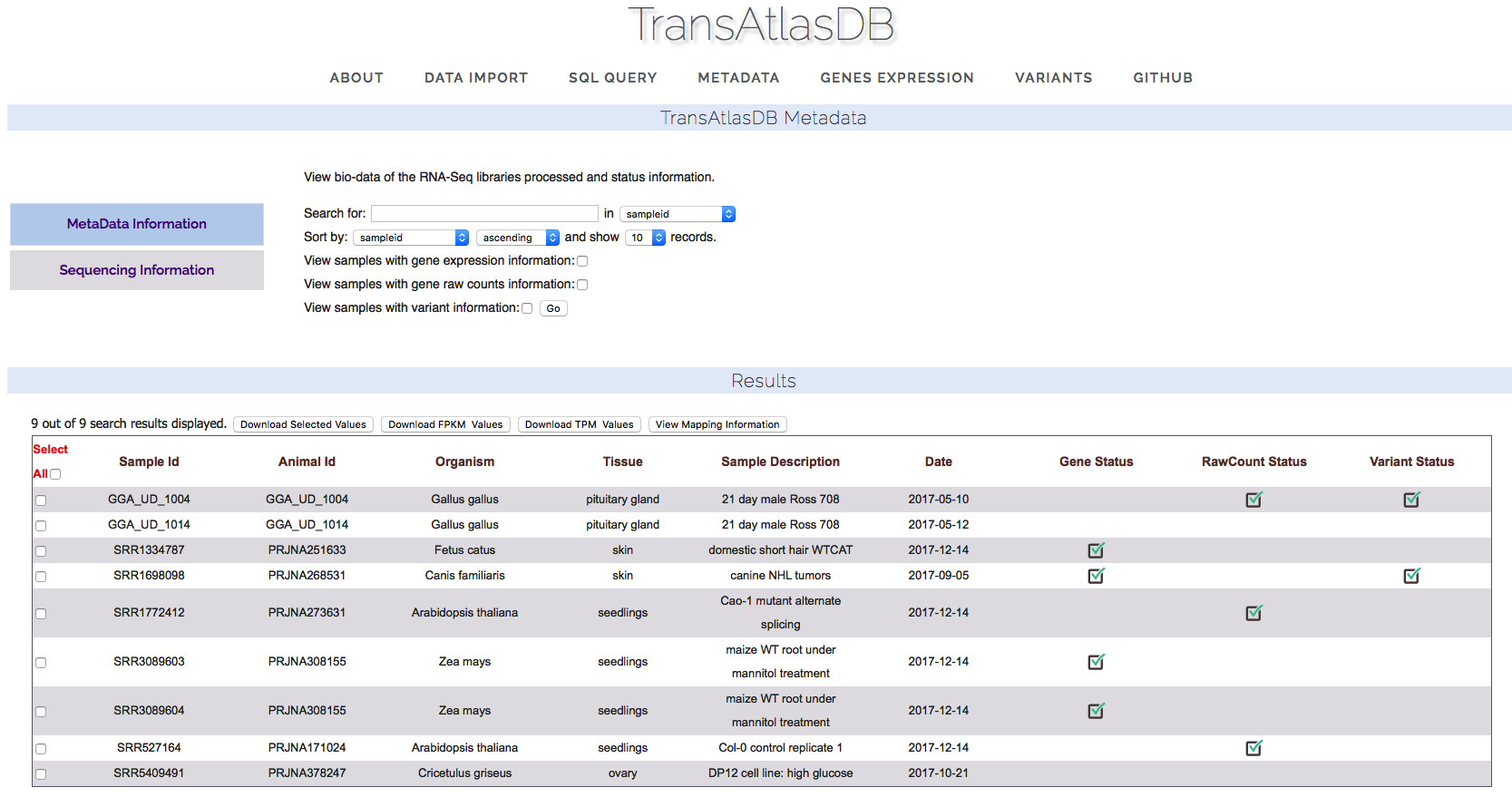
The metadata page displays the samples stored in the database and an overview of each sample storage-status as well as the analysis summary where applicable. The samples can be exported as a tab-delimited file.- Samples Metadata and storage status:
View samples metadata currently in the database based on search criteria.
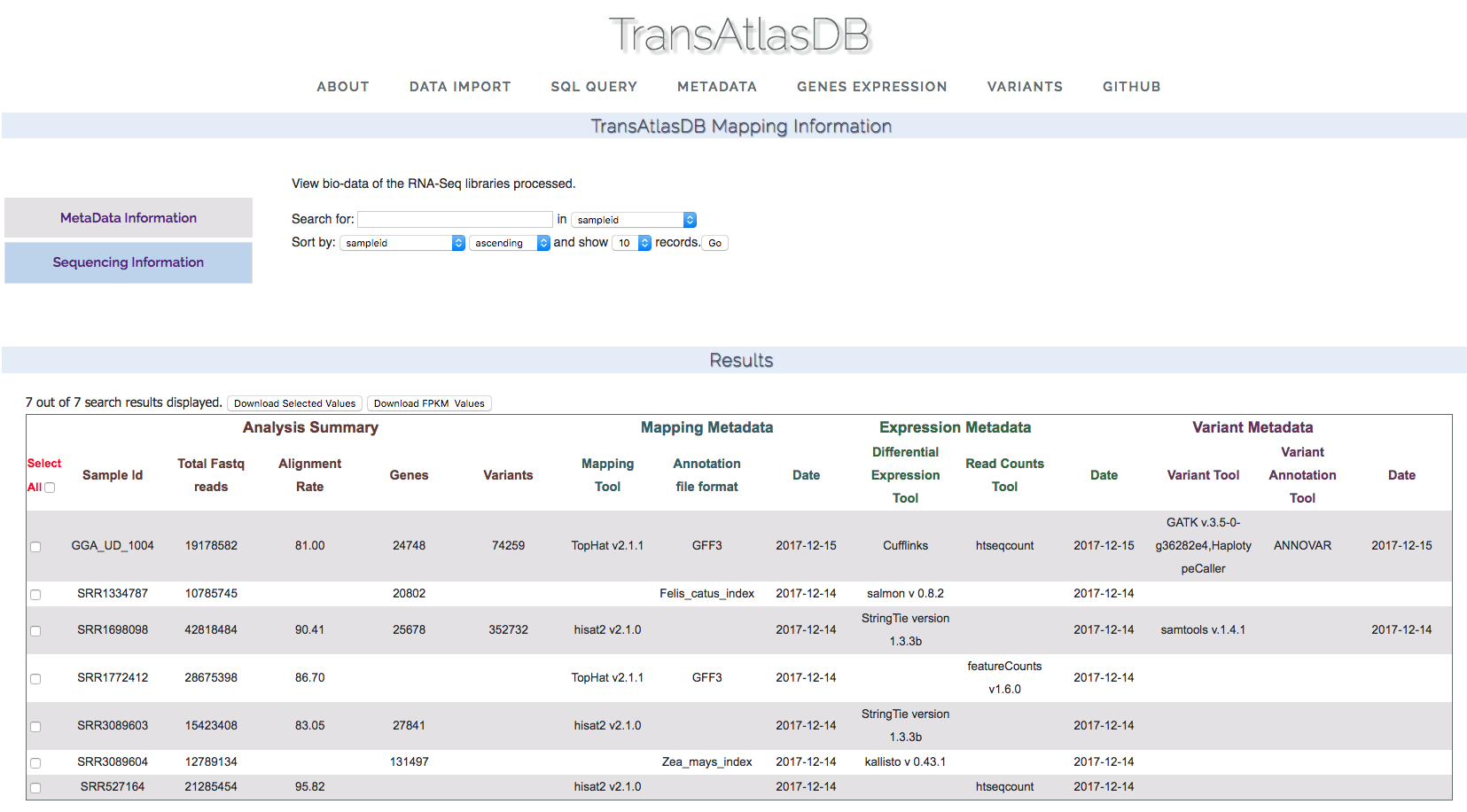
- RNAseq results metadata:
View samples analysis metadata.
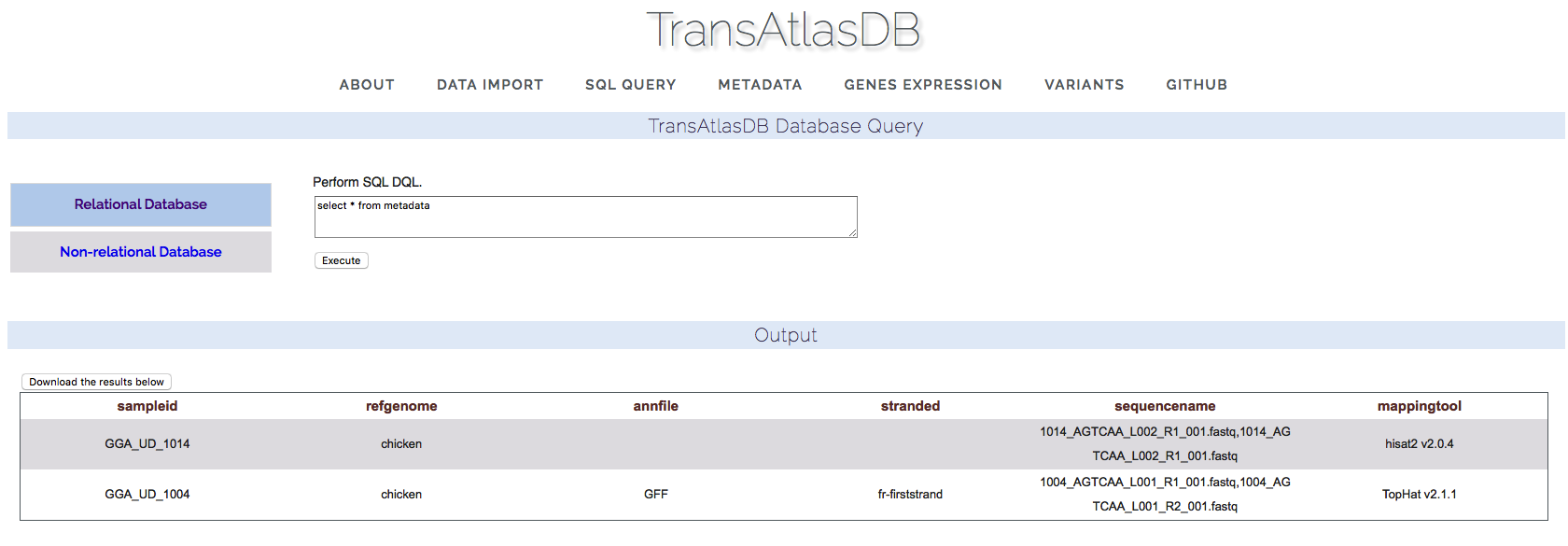
SQL Query
Select statements can be performed on both the MySQL relational database and FastBit NoSQL database. Statements performed will return a table of records based on the select expression, which can be saved as a tab-delimited file.- Relational database.
- Nonrelational database.
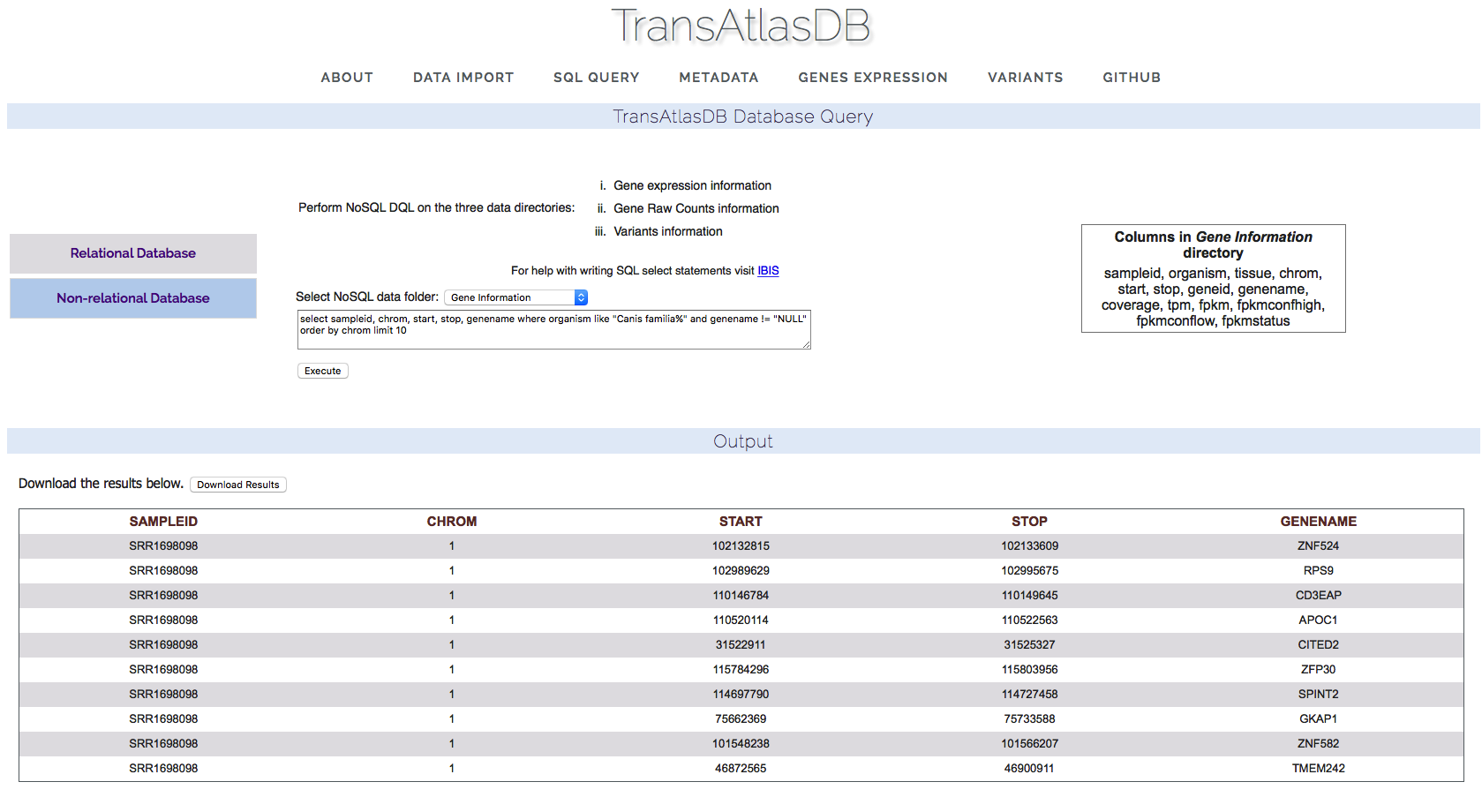
Gene Expression
The gene expression data can be viewed based on the individual sample-gene expression or average expression profiles of multiple genes across all samples and tissues. By specifying one or more genes by their gene symbols, a fuzzy search is performed based on the characters specified. The results can be saved in a tab-delimited file.- Gene Expression summary across all samples.
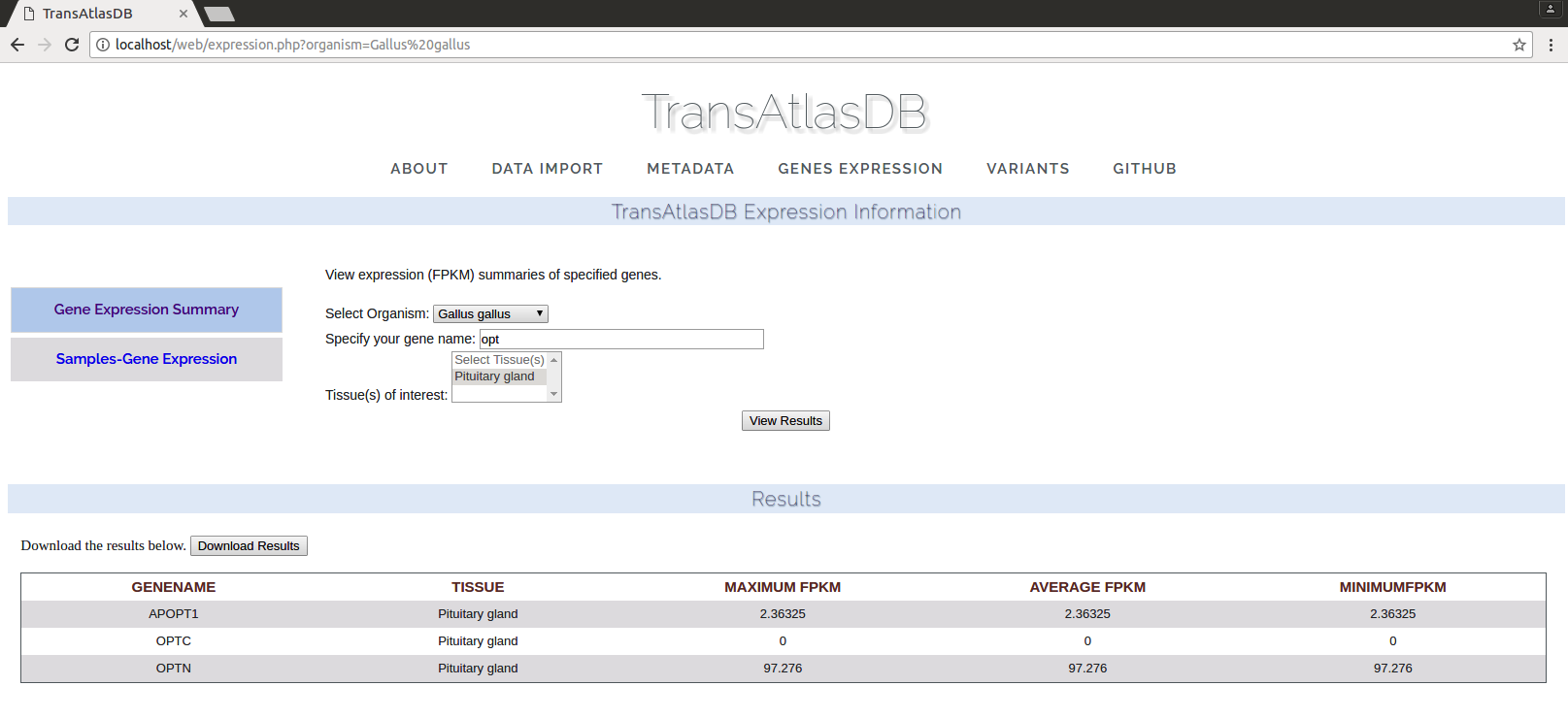
- Gene Expression for each sample
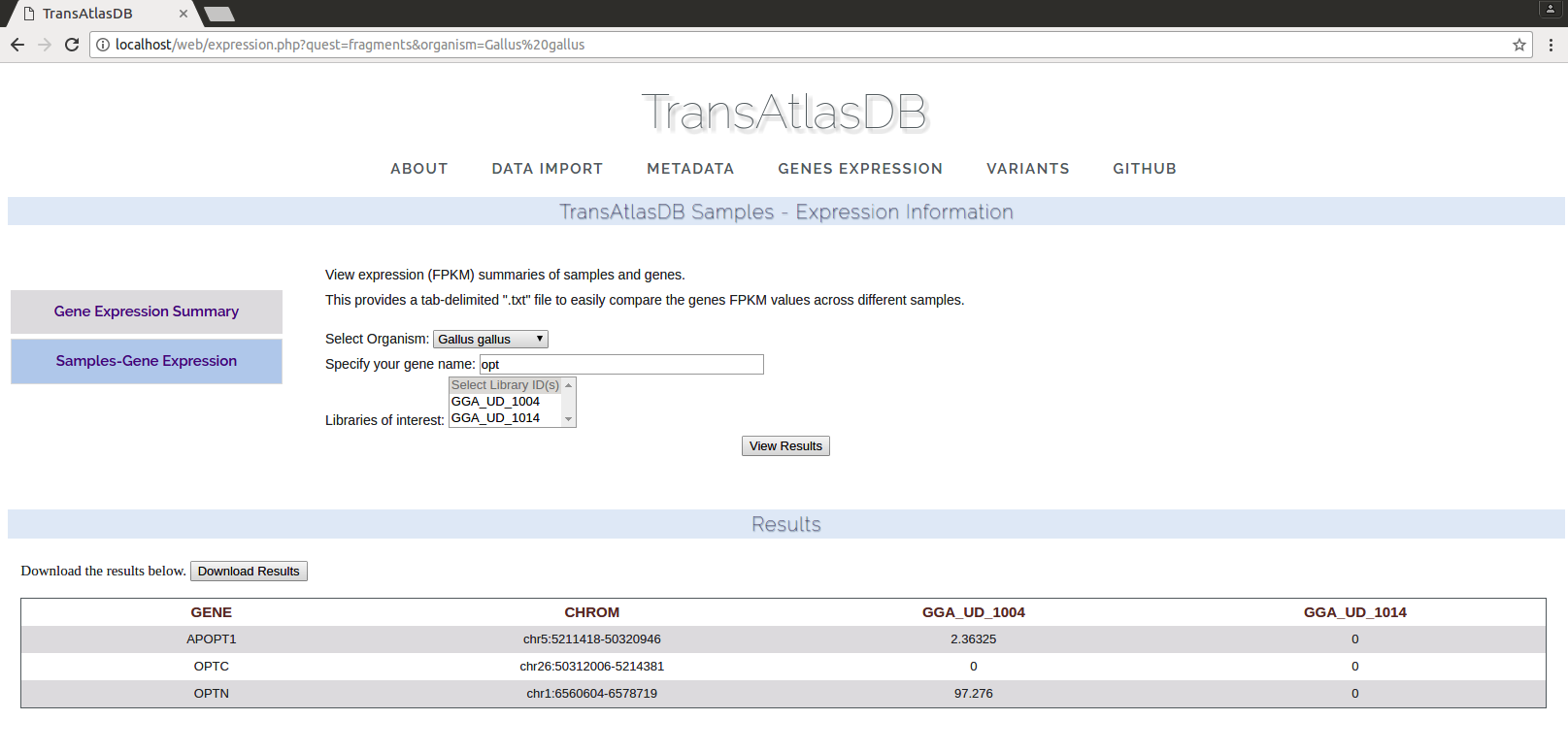
Variants
Variants can be viewed through querying gene symbols or chromosomal regions.- Variant distribution per chromosome and sample
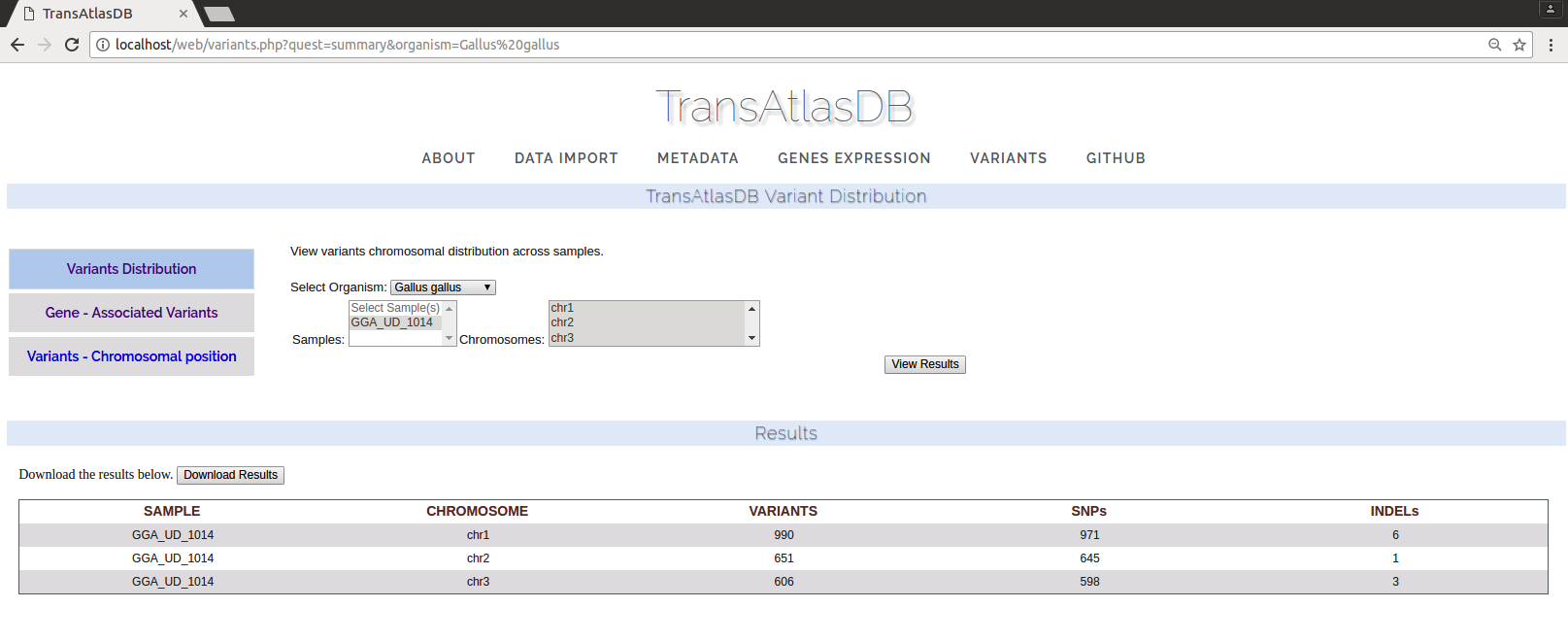
- Variants found in genomic regions of specified genes. One or more genes can be specified and a fuzzy search is performed for each gene queried. The results can be exported as a tab-delimited file.
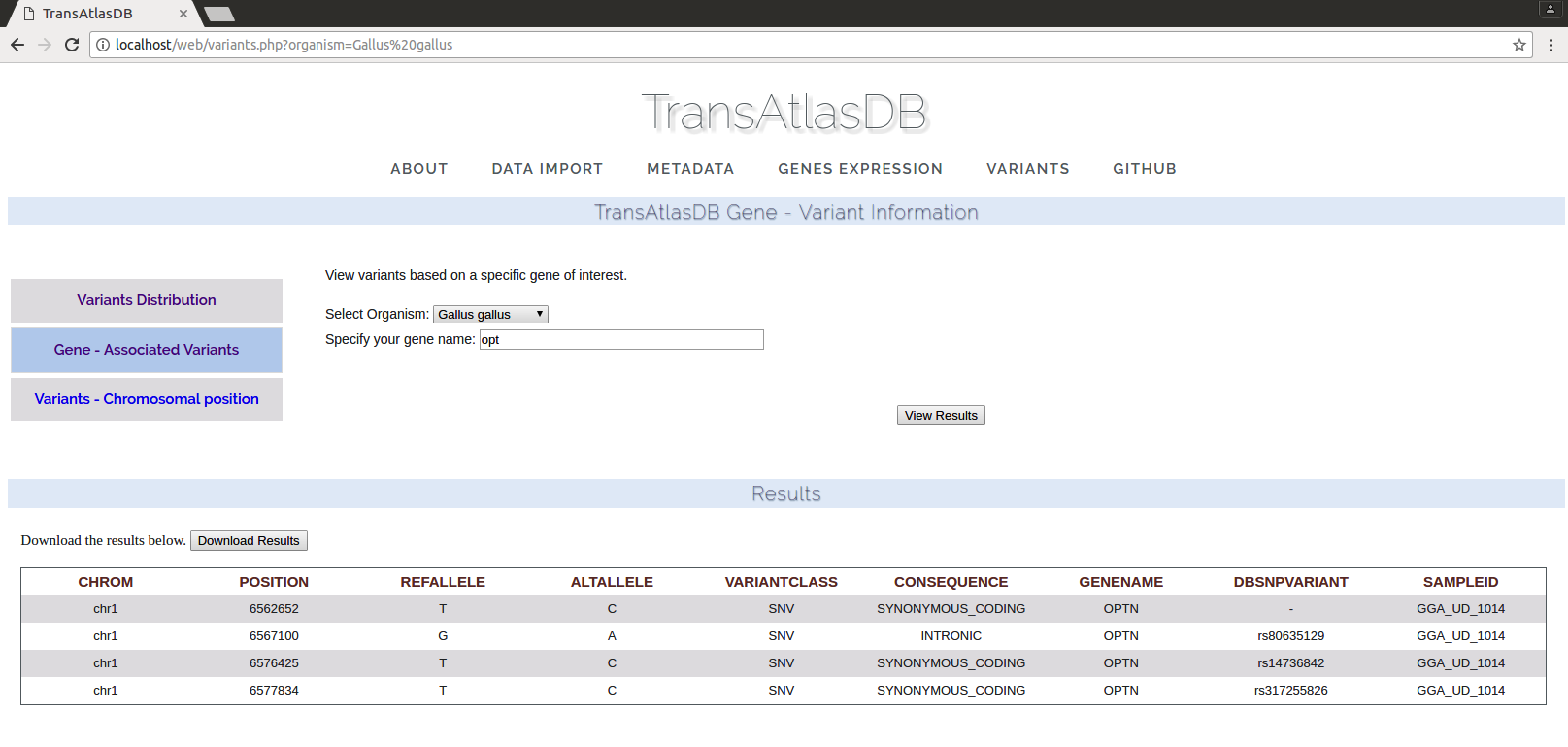
- Variants found based on chromosomal regions specified. These results can be exported as a tab-delimited file or a VCF file for downstream analysis or visualization.

Please click the menu items to navigate through this repository. If you have questions, comments and bug reports, please email me directly.
Thank you very much for your help and support!
Back to Top